Inno-Docking
1. Overview of Inno-Docking
Docking, as a routine technique in structure-based drug design, has been widely used to identify potential Hits in compound libraries, which can help us understand the binding mode between proteins and ligands and estimate their binding affinity. In general, the reliability of a docking program is mainly determined by the efficiency of the conformational search algorithm, which is specialized for generating ligand conformations, and the quality of its scoring function, which is used to calculate binding affinity for protein-ligand binding poses.
A growing number of studies have found that the predicted binding affinity of the scoring functions embedded in docking software does not show a satisfactory correlation with experimentally determined affinities, and even cannot effectively distinguish active and inactive compounds. With the rapid development of computer technology, it was found that scoring functions based on machine learning algorithms can implicitly capture and learn protein-ligands binding characteristics in a non-linear manner, which has been proven to show higher flexibility and better performance than classical scoring functions, which ever in terms of scoring power (the ability to rank binding affinities) , docking power(the ability to distinguish the original binding pose near-native pose from decoys), or screening power (the ability to distinguish the active compound from decoys).
Therefore, the Inno-Docking module not only integrates the classic physical docking program AutoDock Vina, but also integrates our own AI docking program CarsiDock. Compared with physical methods, CarsiDock has significant advantages in docking conformation accuracy. In addition, Inno-Docking also provides complete protein preprocessing, ligand preprocessing, and automatic intelligent docking parameter setting capabilities, and provides detailed data analysis capabilities on the results page.
2. Instruction for Use
On the docking creation task page, we divided the page into two areas according to its functions: the 3D display area, and the parameter setting area. In the 3D display area, you can change the display form of the structure according to your preferences, and long press the left mouse button to rotate the protein 360 degrees, long press the right mouse button to translate the protein, and scroll the mouse wheel to zoom in and out. In addition, according to the docking content, we divided the docking into three steps, namely protein preparation, ligand preparation, and setting docking parameter.
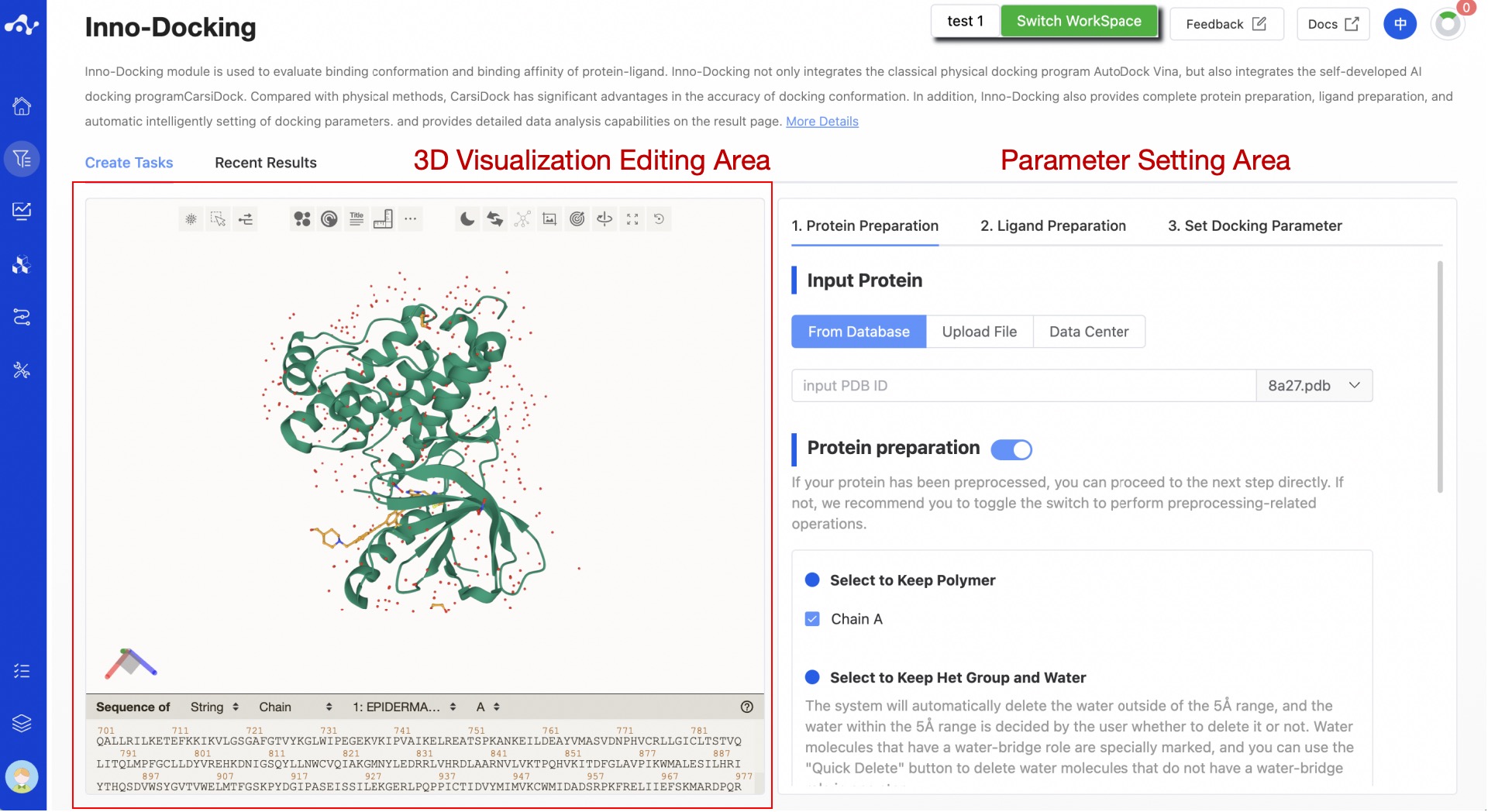
Figure 1. The Inno-Docking calculation page
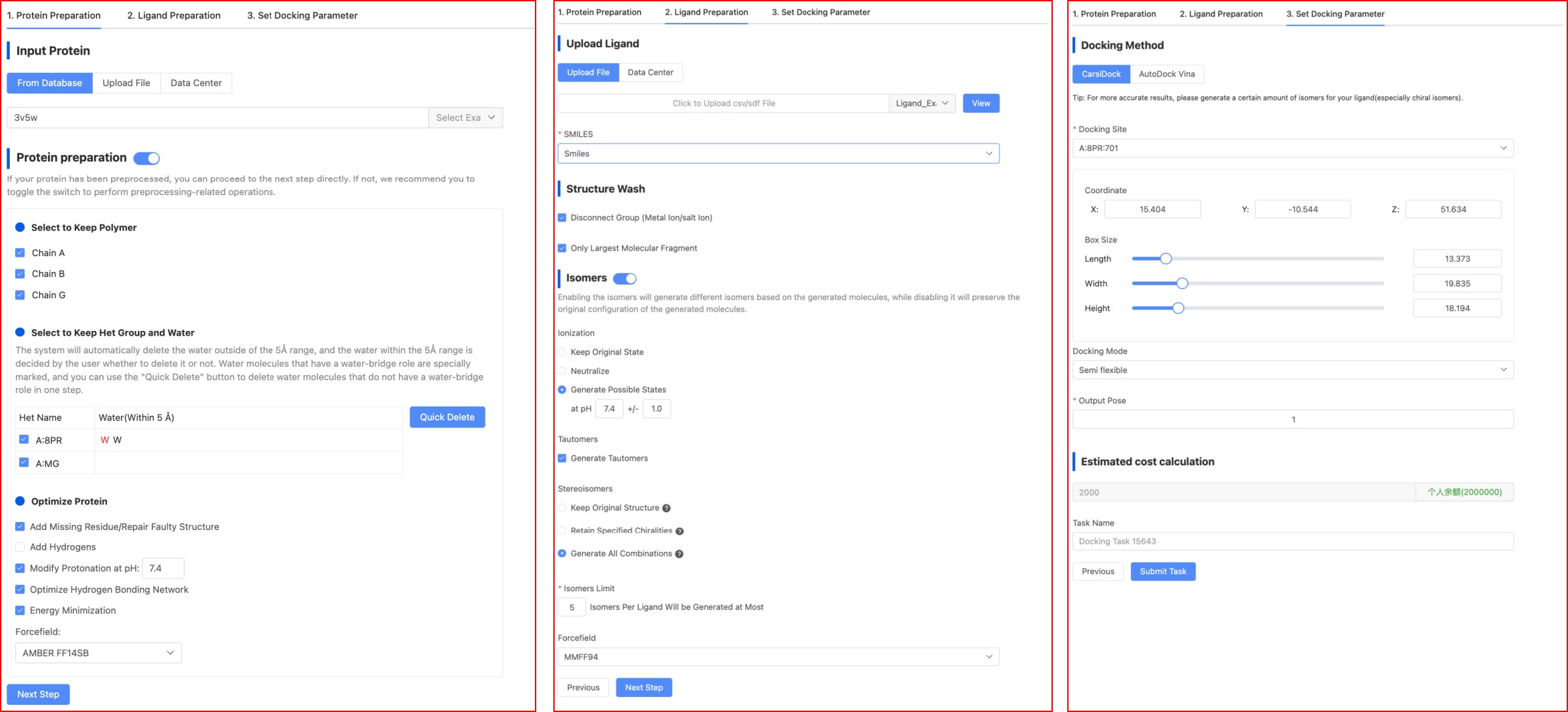
Figure 2. The page of set Docking parameter.
(1) Protein Preparation
According to the conventional protein preparation method, we provide a relatively complete preparation step, including the selection of protein chains, the correction of incorrect structures/repair of missing residues, hydrogenation, local charge calculation, and energy minimization. The panel settings are as shown in Figure 2. The default selections on the page are the current optimal parameters, and users can also choose suitable parameters based on their knowledge.
- Input Protein
The platform offers users two ways to upload protein files: importing from a database and uploading files.
Import from the RCSB PDB database. If the "Database Import" checkbox is checked, when users know the Protein Data Bank (PDB) ID of the protein, they can directly input the 4-digit PDB ID into the text box, and the protein structure will be displayed in the 3D display area on the left.
Upload File. If the "Upload File" checkbox is checked, users can select a local file by clicking the button below. After the file is selected, the file name will be displayed on the button, and the file content will be displayed on the right. The uploaded file only supports .pdb format.
Data Center. If the "Data Center" checkbox is checked, users can select data from the data center by clicking the button below to open a pop-up window and clicking on the file name. After clicking, the pop-up window disappears and the task can be submitted.
Attention! Currently, only CarsiDock supports docking based on water molecules and does not support docking with other cofactors.
- Preparation
The need for protein preparation depends on the type of protein you have uploaded.
If the uploaded protein has already been preprocessed, you can directly click on the next step.
If not, it is suggested that you turn on the switch to do related preprocessing operations.
Select to keep polymer: The system defaults to show all chain information in the uploaded .pdb file, with all chains being selected. When a certain chain is deselected, that chain will not be visible in the protein visualization area.
Select to keep het group and water: This defaults to showing all small molecules and their water molecules within a 5Å range in the pdb file. The system will default to deleting water outside the 5Å range, while water within the 5Å range is up to the user to decide whether to delete. Water molecules that have a water bridge effect have a special mark; you can use the [Quick Delete] button to delete the water that do not have a water bridge effect in one step.
- Protein Optimization
Add missing residues and repair incorrect structures: Optional, selected by default;
Add hydrogen to the protein: Mandatory;
Adjust protonation state: Optional, selected by default, with a pH of 7.4;
Optimize hydrogen network: Optional, selected by default;
Energy minimization: Optional, selected by default, and the chosen force field is AMBER ff14SB.
AMBER ff14SB (recommended). ff14SB is a protein force field parameter set in the AMBER software package, used to describe atomic interactions within biological molecules. It is a particularly applicable force field parameter set for protein systems in the AMBER14 software package, including additional parameters describing interactions between amino acid side chains and important residues in protein folding. AMBER ff14SB has high accuracy and reliability in describing the conformational and dynamical properties of proteins.
AMBER ff15ipq. ff15ipq is an improved protein force field parameter set in the AMBER14 software package, with higher accuracy and reliability compared to AMBER ff14ipq. AMBER ff15ipq includes more polarizable effects and hydrogen bond parameters, allowing for more accurate description of the electronic structure of proteins.
AMBER96. AMBER96 is an early version of the AMBER software package, which has been developed and optimized over many years and now has updated versions such as AMBER14 and AMBER18. However, AMBER96 is still widely used in the field of biomolecular simulation, especially for early research and some classic simulations.
AMBER99SB. AMBER99SB is an improved version of the AMBER99 force field, including additional parameters describing interactions between amino acid side chains and important residues in protein folding. AMBER has high accuracy and reliability in describing the conformational and dynamical properties of proteins.
CHARMM36. CHARMM36 has high accuracy and reliability in describing the conformational and dynamical properties of proteins. It is widely used in the study of protein-protein and protein-ligand interactions, and is one of the commonly used force field parameter sets in the field of biomolecular simulation.
(2) Ligand Preparation
The platform provides routine small molecule preparation methods, including removing unconnected groups (such as metal ions, salt ions), retaining the largest molecular fragments, generating isomers (ionic states, isomers, stereoisomers) and hydrogenation, and energy minimization. The panel settings for ligand preparation on our platform are as shown in Figure 2. The default selections on the page are the current optimal parameters, and users can also choose suitable parameters based on their knowledge. The parameters here are consistent with those in the "Ligand Preprocessing" module.
- Input ligand
The platform currently supports upload ligand through file upload only.
Upload files. Check the "Upload File" box, and you can select a local file by clicking the button below. After the file is selected, the file name will be displayed on the button, and the file content will be displayed on the right. The uploaded files should be in .pdb format.
Data Center. Check the "Data Center" box, and a pop-up window will appear when you click the button below. Click on the file name to select data from the data center. After you click, the pop-up window will disappear and you can submit the task.
- Isomers
The current platform enumerates the uploaded molecules by default (the switch status is "on") in order to generate more isomers, including ionic isomers, tautomers, and stereoisomers. When the switch status is "off", the system will not do other processing on the molecules and will retain the original conformation of the molecules.
Ionic isomers. It generates possible ionization states by adjusting the pH value range;
Tautomers. It generates possible isomers based on the ionization state;
Stereoisomers. It generates possible isomers based on the chiral features of the molecule, or it can retain the original state.
Number of isomers. The platform defaults to output a maximum of 5 isomers. Users can adjust according to their needs.
- Forcefield
MMFF94. MMFF is an abbreviation for Merck Molecular Force Field, a professional small molecule force field. It's the second-generation molecular force field developed by Hagler and is one of the most accurate force fields currently available.
UFF. UFF is an acronym for Universal Force Field. It is a general force field that covers the entire periodic table of elements. Its accuracy in calculating structures and binding energies is average, and it is used only when a suitable force field cannot be found.
(3) Set Docking Parameter
In this step, users need to first select a docking method, and then set related parameters based on the selected docking method.
- Choose the Docking Method
CarsiDock is an AI docking algorithm independently developed by CarbonSilicon AI, with fast speed and high precision. AutoDock Vina, on the other hand, is open source software. We have optimized it to improve both the speed and precision compared to the original version.
- Docking site
Both CarsiDock and AutoDock Vina support two ways to define docking sites, namely selecting a ligand in the complex and customizing the docking site. The default coordinates are randomly assigned by the system, and the default length, width, and height are 15Å. These parameters represent the location and size of the binding pocket.
Choose the ligand in the complex as the docking site. This option only applies when the complex naturally contains a ligand. When there are multiple ligands in the complex, the system defaults to display the ligand with the largest molecular weight. Users can switch to other ligands in the drop-down box. According to the selected ligand, the system will calculate the geometric center (XYZ coordinates) based on the size of the ligand and add 5Å to the edge of the ligand.
Customize the docking site. In custom mode, users can directly place the mouse in the 3D display area. After you click on a residue, the corresponding coordinate information of the residue will be displayed on the parameter panel, and the system will also give a default length, width, and height of 15Å. However, the specific size of the pocket needs to be set by the user after understanding the protein pocket. If the pocket is too small, the calculation result will be inaccurate, and if the pocket is too large, the computation time will increase.
- Docking Mode
Both CarsiDock and AutoDock Vina only support semi-flexible docking methods, that is, allowing the ligand to vary within a certain range while keeping the conformation of the receptor unchanged.
- Output Conformation
By default, one docking conformation is output for each molecule. Users can adjust the number of outputs via the input box, up to 100.
(4) Running Sataus and View Results
After the task is submitted, the page will automatically jump to the "Recent Results" subpage of the current page. Here you can view the task running status of the current module (progress bar), and you can also view all running tasks of all modules in the "Running" dropdown box in the upper right corner. When the data volume is large, the system will calculate in batches, so as long as a batch of data is calculated (while the entire task is still running), you can click the "Result Details" button to enter the result page and view the prediction result list of the currently completed calculations (molecules that have not completed the calculation will not be displayed temporarily). You can also refresh the current page to get the latest completed data.
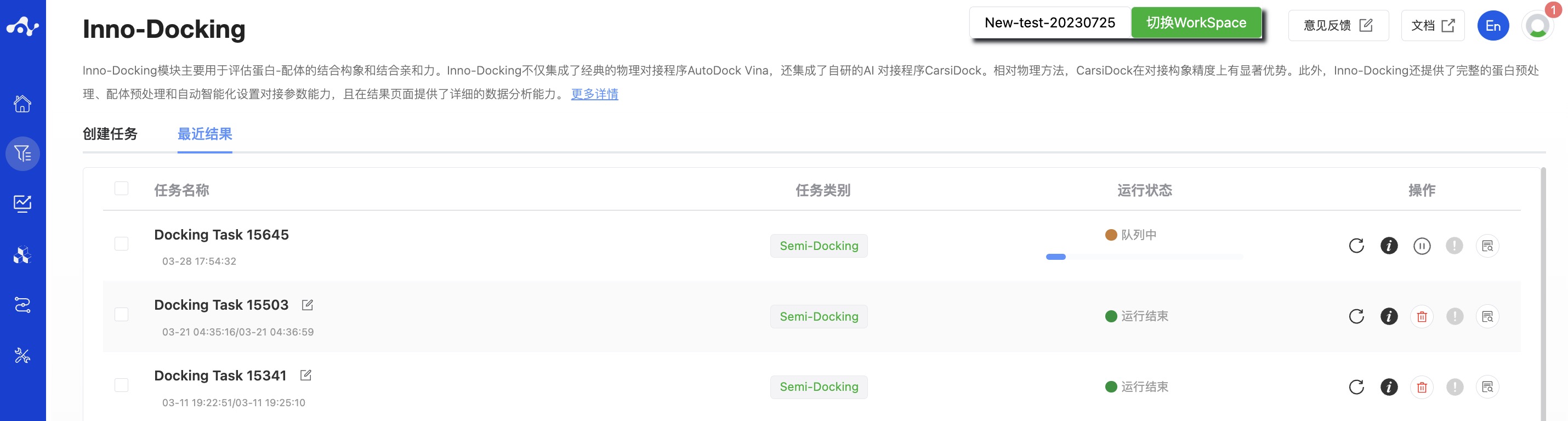
Figure 3. View results
3. Result Analysis
The results page is composed of the Summary at the top, the molecule list on the left, and the protein visualization area on the right. By default, the result details area on the left displays the list page, but you can switch to the grid page. The grid page provides a simple page for viewing molecular structures, while the list page provides detailed calculation results, making it easier for you to analyze the current data. The protein visualization area is a fixed content, no matter what sub-page is on the left, the protein visualization area will display the protein-ligand docking mode. Users can quickly browse the interactions between molecules through the "Previous" and "Next" buttons at the bottom right.
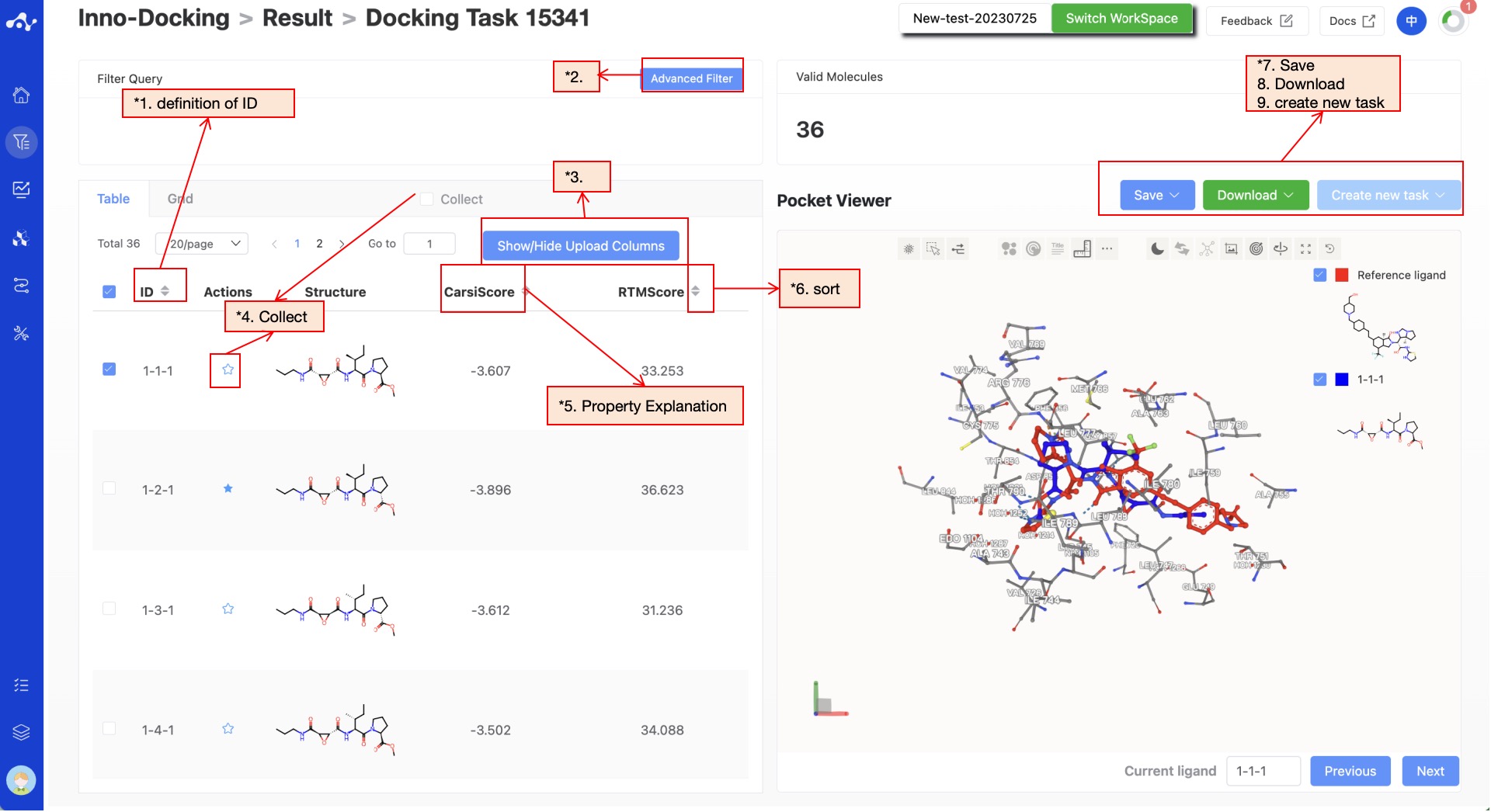
Figure 4. Results Page Function Distribution
(1) Meaning of ID in the table
The ID is assigned according to the molecule order in the original file. If the task does not generate isomers, the ID will be continuous values from 1 to N; if the task chooses to generate isomers, then the ID will be a combination of X-Y-Z. X represents the order of molecules, Y represents the number of isomers of the molecule, and Z represents the number of conformations output. The smaller the Y value, the higher the likelihood of this isomer existing.
(2) Advanced Filtering
Advanced filtering provides range filtering, which can further filter out molecules within a specified range of certain properties to exclude molecules that do not meet the expected results. After advanced filtering, only molecules that meet the filter conditions will be displayed on the page.
(3) Show/Hide Upload Column
The default result list does not display information in the uploaded file, so it is unselected in the left control bar. When you don't want to display this property, deselect it, and the result list on the left will show in real time based on the selection in the control bar. At the top, there are also two shortcuts "Select All" and "Deselect", which are convenient for users to quickly select.
(4) Favorites
This feature is mainly used to help users mark their favorite molecules. When you click to add a molecule to your favorites, the icon will be highlighted, meaning that the molecule is marked as a favorite. After clicking the favorite checkbox, the page will only display the favorited molecules. After favoriting, you can click the chart again to cancel the favorite.
(5) Property Explanation
Hover the mouse over the name of each property to view the interpretation of the corresponding attribute.
(6) Sorting
Click the property name in the result list to reorder. For example, F(20%), click once for ascending order, click again for descending order, and click a third time to restore the original order.
(7) Save
Click "Save", and the system will pop up a dropdown box for you to choose the file format to save (currently only supports .csv/.sdf). Once you have determined the style of the file to save, save the corresponding data to the data center as a sdf or csv file. The saved content is the molecules of the effective number displayed on the page, which are usually obtained according to your show/hide column conditions, advanced filtering conditions, favorites, or dislikes.
(8) Download
Click "Download", and the system will pop up a dropdown box for you to choose the file format to download (currently only supports .csv/.sdf). After determining the style of the file to download, the system will download the corresponding data to your local device as a sdf or csv file. The content downloaded is consistent with the save method, which also downloads the molecules of the effective number displayed on the page, which are usually obtained based on your show/hide column conditions, advanced filtering conditions, favorites, or dislikes.
(9) Create New Task
The prerequisite for creating a new task is to first save the data into a file. Before the save operation is performed, this button is disabled. As soon as the new file is saved based on the results, this button is enabled. When you click this button, the system will pop up a dropdown box for you to select the module to be calculated. After clicking, the page will immediately open a new tab and will take your saved dataset with it. After adjusting the parameters, you can submit a new task.
4. Introduction to the Related Algorithm-CarsiDock
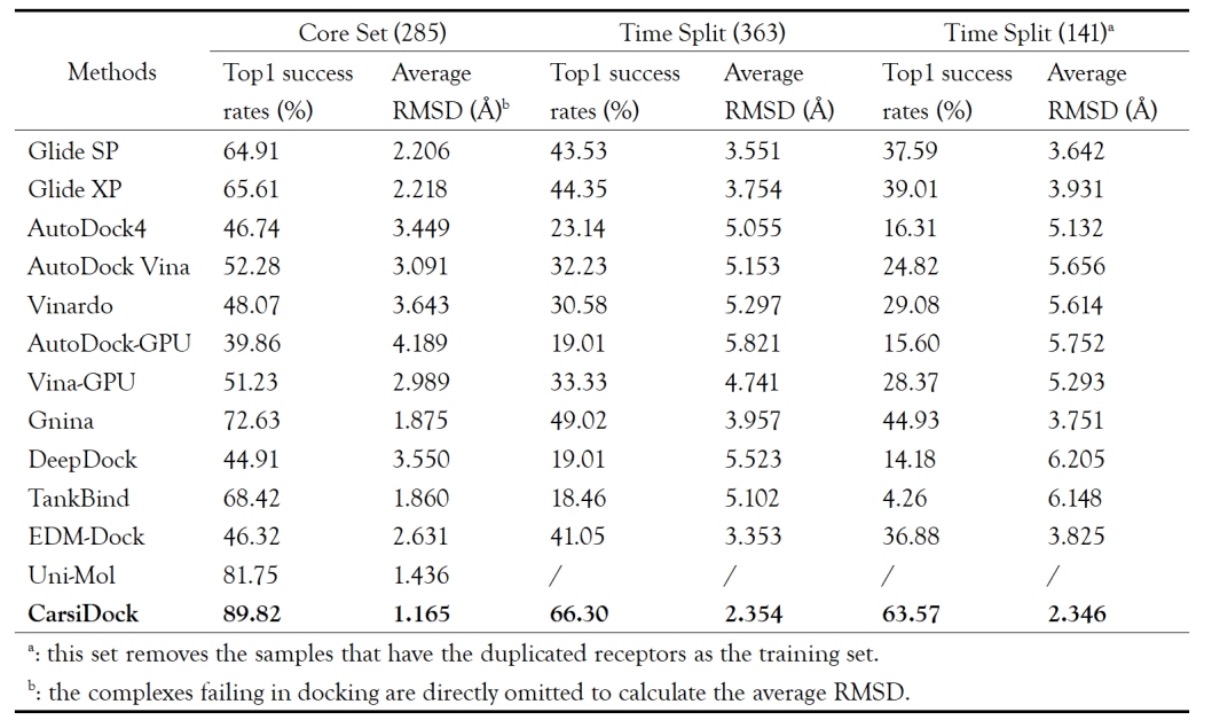
CarsiDock is a semi-flexible AI docking method developed by CarbonSilicon AI, which innovatively uses a rigid docking-guided self-distillation method and is pre-trained on large-scale physical simulation data. The CarsiDock method is inspired by pre-training methods like BERT/ChatGPT and effectively combines physical docking with AI docking methods: firstly, it uses AI methods to predict the binding mode of proteins and small molecules, then uses gradient descent to quickly obtain docking conformations. Furthermore, it uses physical docking methods to generate a large amount of docking data. This physical simulation data includes important information such as the structure of molecules, modes of action, and binding affinity. Based on this data, CarsiDock conducts pre-training, which provides a well-abstracted foundational model for AI docking. Crystal data refinement is then applied to further enhance the accuracy of AI predictions. The results show that CarsiDock can not only ensure the topological reliability of binding postures but can also successfully reproduce key interactions in crystalline structures, highlighting its superior applicability and excellent docking and screening capabilities.
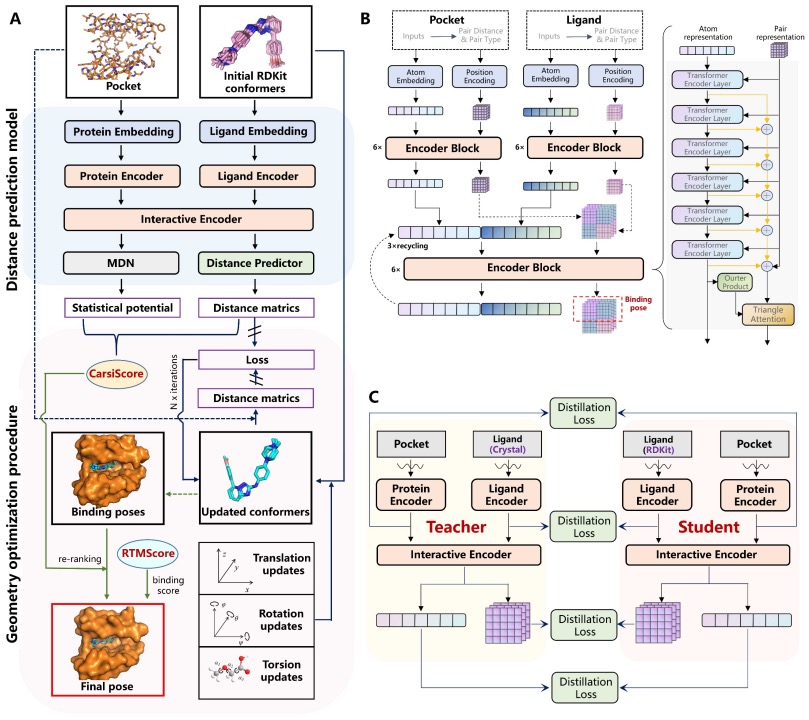
Figure 5. The framework of CarsiDock Model
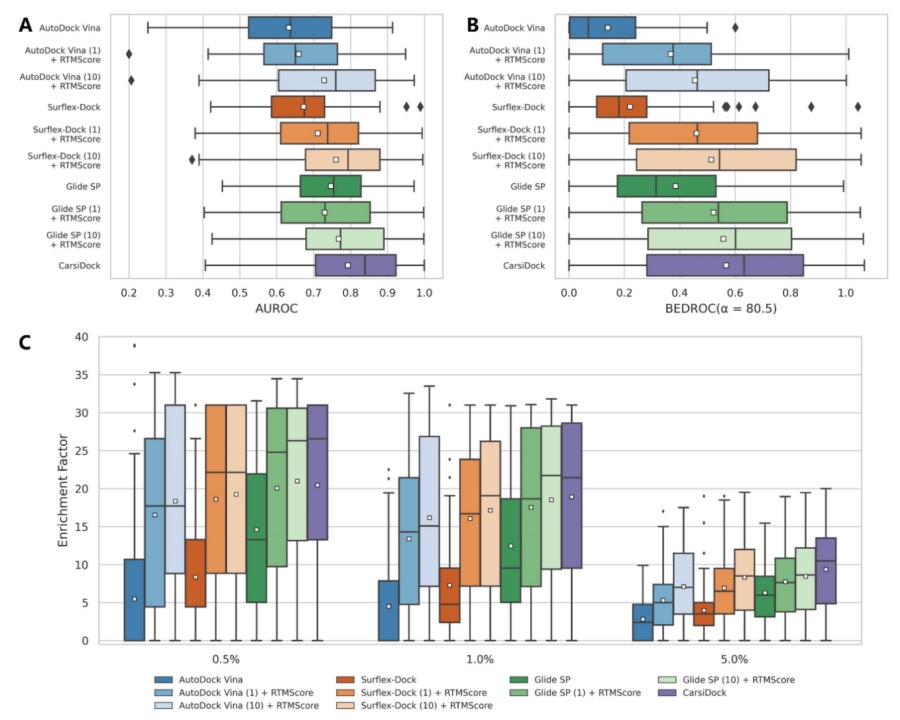
Figure 6. Screening Performance of Various Methods on the DEKOIS2.0 Dataset.
Table 1. Comparison of Top1 Success Rate and Average RMSD on the PDBBind CoreSet and Time-Split Dataset.
5. Related Literature
【1】CarsiDock: a deep learning paradigm for accurate protein-ligand docking and screening based on large-scale pre-training. In Submit
