Inno-Rescoring
1. Overview of Inno-Rescoring
The Inno-Rescoring module offers excellent scoring functions based on machine learning algorithms. To make it easier for users to directly apply our rescoring functions, the Rescoring module allows users to upload related files for rescoring after local docking. Users can choose these rescoring functions to reevaluate the binding affinity of protein-ligand binding poses.
2. Instructions for Use
Users only need to complete four steps to finish the calculation: upload the preprocessed protein, upload the post-docking ligand, name the task (optional), and submit the task (mandatory).
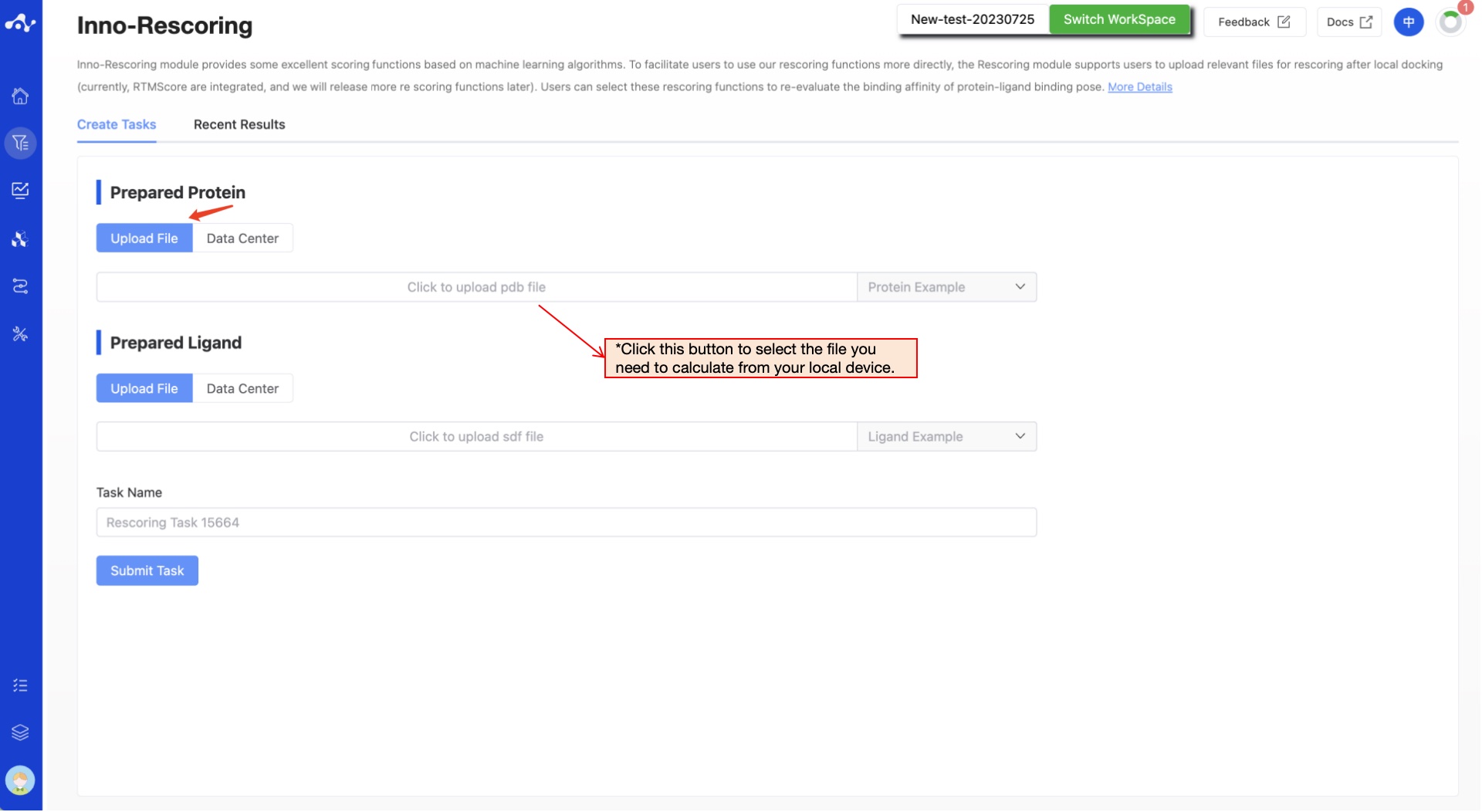
Figure 1. Upload prepared protein and ligand - Upload file/Data Center
(1) Upload Prepared Protein
The platform provides two ways to upload preprocessed proteins: Upload File and Data Center.
- Upload File
Check the "Upload File" box, and you can select a local file by clicking the button below. After the file is selected, the file name will be displayed on the button, and the file content will be displayed on the right.
- Currently supported file format: .pdb
- Data Center
Check the "Data Center" box, and a pop-up window will appear when you click the button below. Click on the file name to select data from the data center. After you click, the pop-up window will disappear and you can submit the task.
(2) Upload Prepared Ligand
The platform provides two ways to upload preprocessed proteins: Upload File and Data Center.
- Upload File
Check the "Upload File" box, and you can select a local file by clicking the button below. After the file is selected, the file name will be displayed on the button, and the file content will be displayed on the right.
- Currently supported file format: .sdf
- Data Center
Check the "Data Center" box, and a pop-up window will appear when you click the button below. Click on the file name to select data from the data center. After you click, the pop-up window will disappear and you can submit the task.
After all parameters are set, name the task and click submit to complete the task submission.
(3) Running Sataus and View Results
After the task is submitted, the page will automatically jump to the "Recent Results" subpage of the current page. Here you can view the task running status of the current module (progress bar), and you can also view all running tasks of all modules in the "Running" dropdown box in the upper right corner. When the data volume is large, the system will calculate in batches, so as long as a batch of data is calculated (while the entire task is still running), you can click the "Result Details" button to enter the result page and view the prediction result list of the currently completed calculations (molecules that have not completed the calculation will not be displayed temporarily). You can also refresh the current page to get the latest completed data.
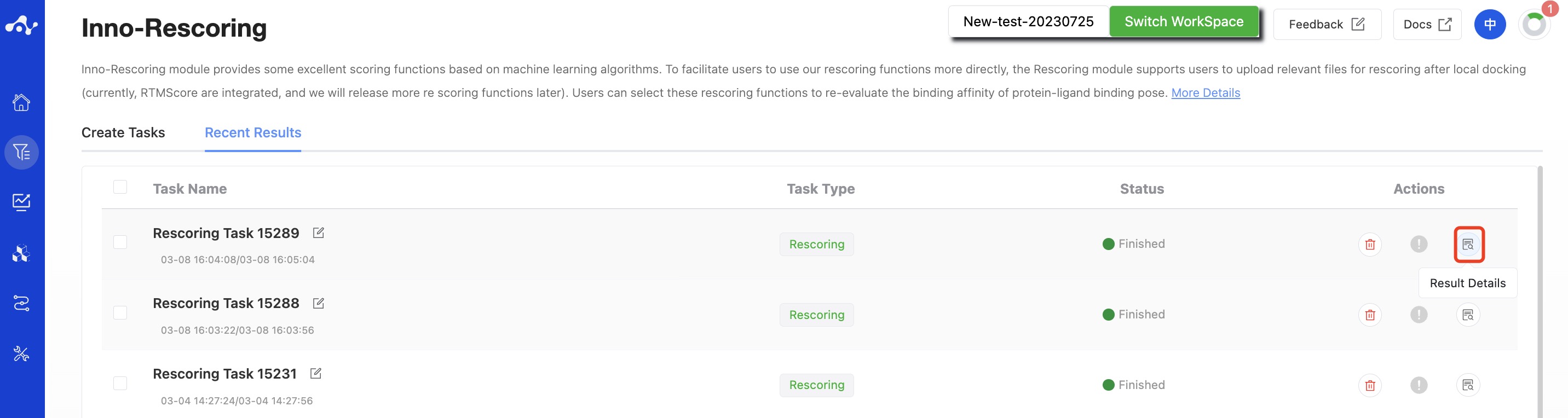
Figure 2. View Results
3. Results Analysis
The results page consists of a top Filter area, a Summary, and a central Results Details area. By default, the Results Details area displays a results list (you can also switch to card view). At this time, you can view all the scores and sort these scores as needed.
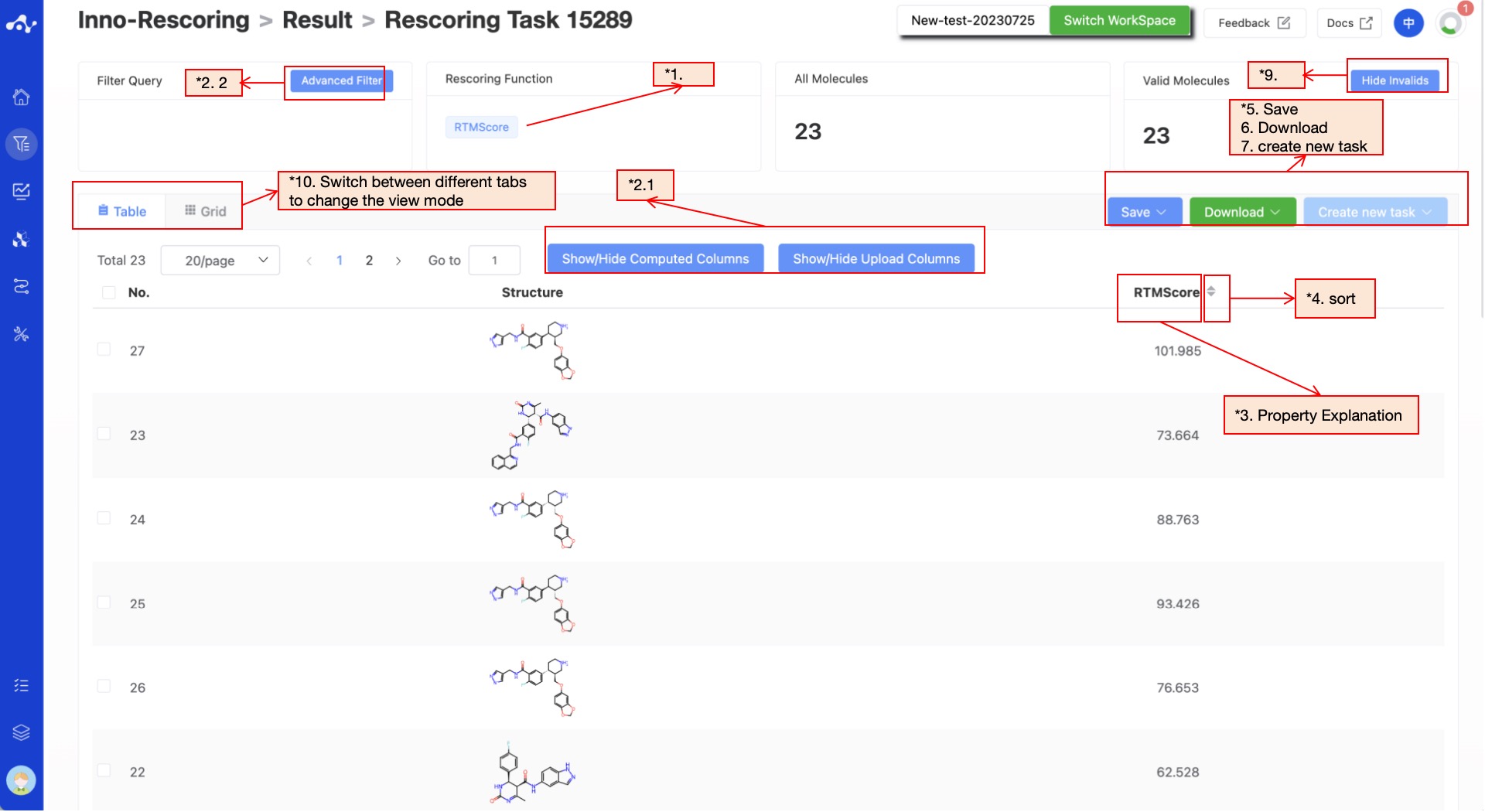
Figure 3. Results Page Function Distribution
(1) Rescoring Function
The Rescoring Function is RTMScore.
(2) Filter
The platform provides general filter and advanced filter to meet the needs of users.
- General filter
General filters can show/hide properties. The default result list shows all the computed properties, and the control column on the left is in the selected state. When you do not want to display the property, just uncheck the property, and the result list on the left will be displayed in real time according to the selection in the control bar. Two shortcut keys "Select All" and "Select None" are also provided at the top, which is convenient for users to quickly select.
- Advanced filter
On the basis of general filter, advanced filter provides range screening, which can further screen out molecules within a specified range of a property to exclude molecules that do not meet the expected results.
(3) Attribute Interpretation
Hover your mouse over the name of each property to see the corresponding explanation.
(4) Sorting
Click the property name in the result list to re-sort, such as F(20%), click once to ascending order, click again to descending order, click again to restore the original sort.
(5) Save
Click "Save", and the system will pop up a dropdown box for you to choose the file format to save (currently only supports .csv/.sdf). Once you have determined the style of the file to save, save the corresponding data to the data center as a sdf or csv file. The saved content is the molecules of the effective number displayed on the page, which are usually obtained according to your show/hide column conditions, advanced filtering conditions, favorites, or dislikes.
(6) Download
Click "Download", and the system will pop up a dropdown box for you to choose the file format to download (currently only supports .csv/.sdf). After determining the style of the file to download, the system will download the corresponding data to your local device as a sdf or csv file. The content downloaded is consistent with the save method, which also downloads the molecules of the effective number displayed on the page, which are usually obtained based on your show/hide column conditions, advanced filtering conditions, favorites, or dislikes.
(7) Create New Task
The prerequisite for creating a new task is to first save the data into a file. Before the save operation is performed, this button is disabled. As soon as the new file is saved based on the results, this button is enabled. When you click this button, the system will pop up a dropdown box for you to select the module to be calculated. After clicking, the page will immediately open a new tab and will take your saved dataset with it. After adjusting the parameters, you can submit a new task.
(8) Hide Invalid Molecules
For SMILES errors or molecules that cannot be resolved in the back-end, the algorithm cannot perform the correct calculation, in which case the task is unaffected, but the molecule is defined as invalid. Users can use the "Hide invalid molecules" button to quickly filter out the part of the molecule.
(9) Card Subpage
In the card page, we provide a more concise way to view the results, you can only see the structure of the molecule and its rescoring value RTMScore.
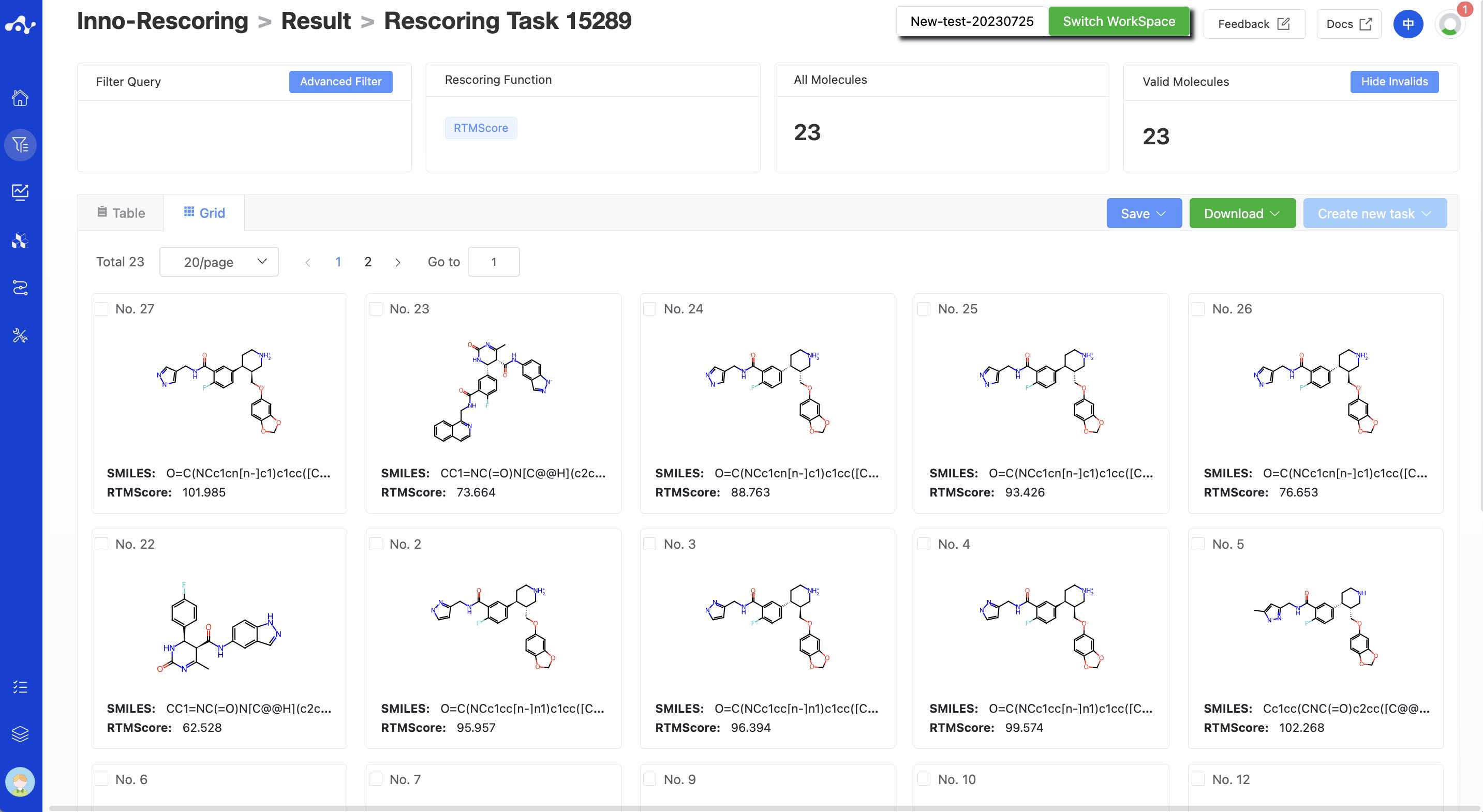
Figure 4. Grid page of the results
4. Introduction to the Related Algorithm
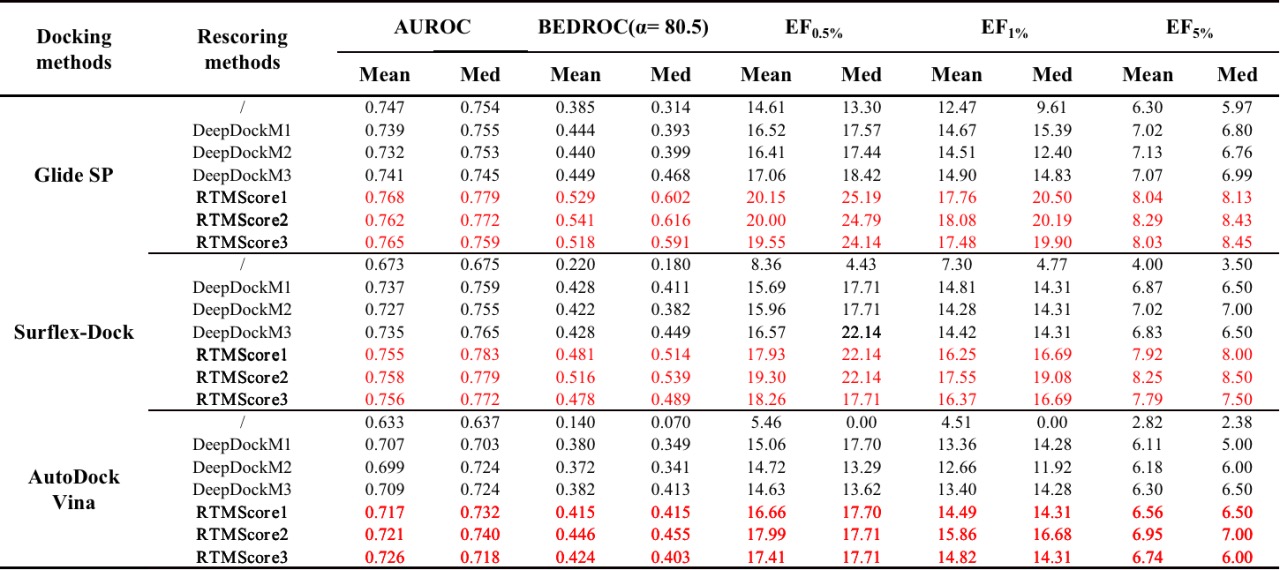
RTMScore is a novel scoring function for the prediction of protein-ligand binding affinity, which introducing graph transformer for the feature extraction of proteins and ligands, and using MDN(Mixture Density Network) to obtain the probability density distribution of the distance between each residue and each ligand atom, and finally converted the density model into the statistical potential to evaluate the binding affinity of protein-ligand. Results proved that RTMScore can outperform almost all the other state-of-the-art scoring functions in terms of both the docking and screening powers on CASF-2016 benchmark. In terms of docking power, RTMScore can achieve an average top1 docking success rate of 97.3% and 93.4%, respectively, with or without the consideration of the native poses(DeepDock and PIGNet wewre only 87% in the absence of the native poses) . As for screening power, RTMScore can yield an average top1 success rate of 66.7% and a 1% enrichment factor of 28.00 (55.4% and 19.60 for DeepDock and 43.9% and 16.41 for PIGNet, respectively). Further screening power evaluation on DEKOIS2.0 was performed, results showed that the enrichment ability of RTMScore significantly outperformed to the similar model architecture DeepDockM and classical method Glide SP. For details, please refer to the reference [1].
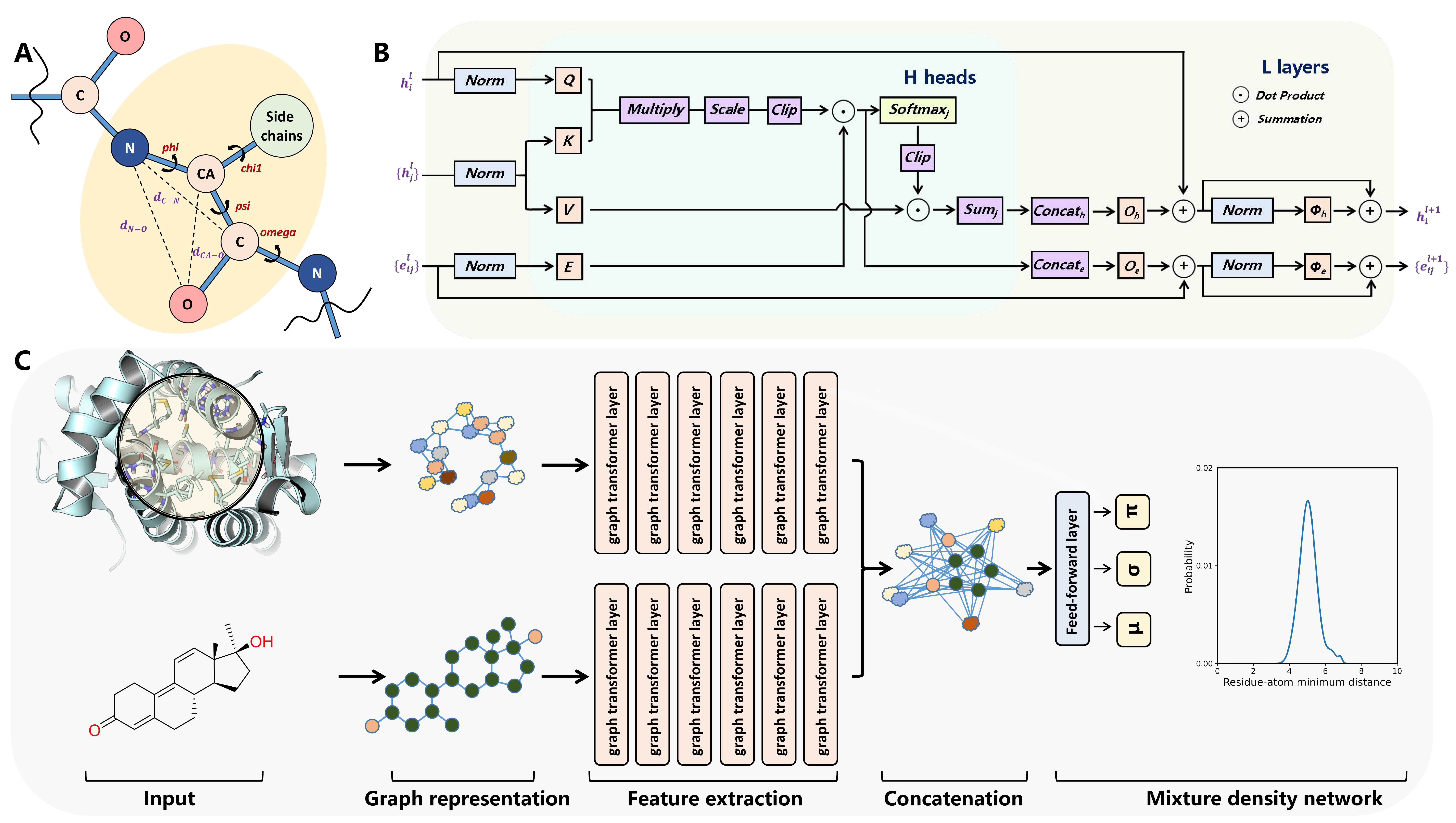
Figure 5. The framework of RTMScore model.
Table 1. Comparison of RTMScore with other state-of-the-art methods in terms of docking power and forward screening power on CASF-2016 benchmark.
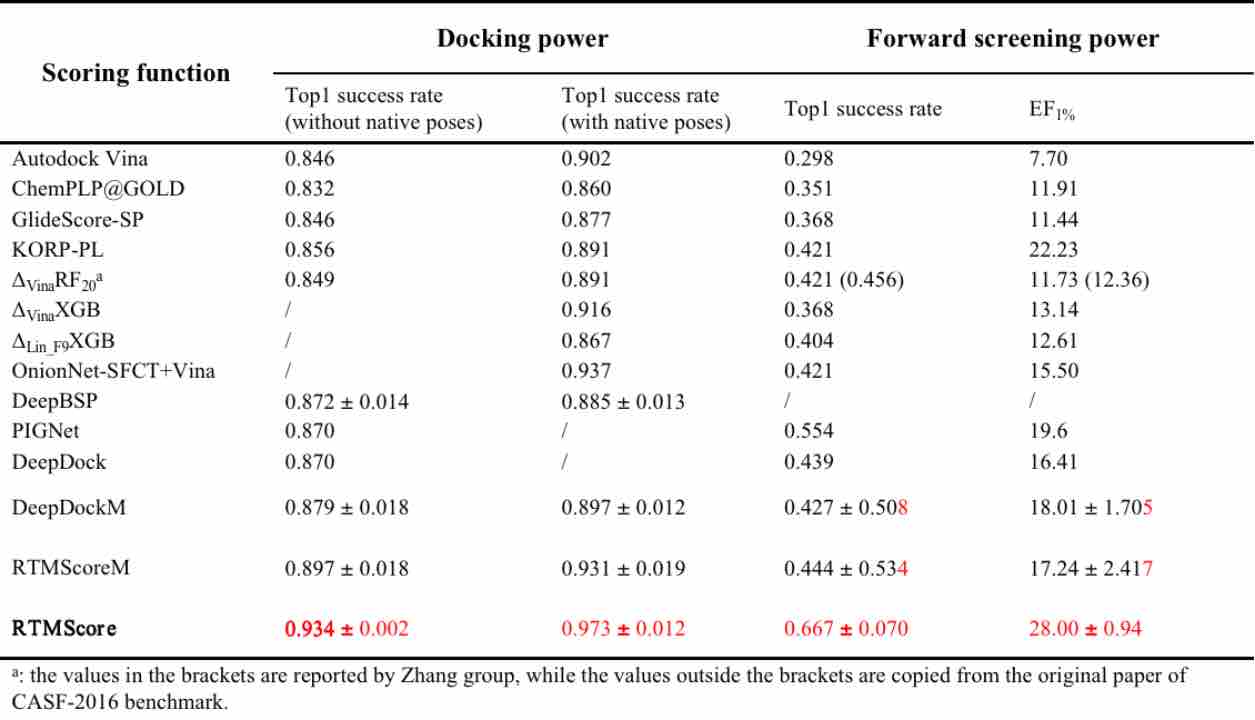
Table 2. Screening powers of scoring functions on DEKOIS2.0 dataset.
5. Related Literature
[1] Boosting Protein-Ligand Binding Pose Prediction and Virtual Screening Based on Residue−Atom Distance Likelihood Potential and Graph Transformer. Shen C, Zhang X, Deng Y, et al. J Med Chem. 2022. doi:10.1021/acs.jmedchem.2c00991
