Inno-QSAR
1. Overview of Inno-QSAR
Quantitative Structure activity Relationship (QSAR) is a method to study the relationship between chemical structure, physical and chemical properties and biological activity of compounds, so as to establish a quantitative (qualitative) prediction model. With the rapid development of computer technology, it has now developed into QSAR models based on various machine learning (ML) algorithms, and ML algorithms such as Random Forest (RF), eXtreme Gradient Boosting (XGBoost) have gradually replaced the primary Hansch method and Free wilson method. By using these ML/DL algorithms to learn the relationship between known bioactive compounds and their corresponding structures or physical properties, a QSAR classification or regression model can be established to effectively differentiate actives from inactives or predict the biological activity of new compounds.
For a model, the key point is the performance of the model. There are three main factors that affect the performance of the model, namely, dataset, descriptor and algorithm.
Dataset. Dataset is the cornerstone of a model, which directly determines the reliability and practicability of the final model. The dataset with diversity structures and rich data, which means that the model has wide application threshold and strong practicability. Therefore, users should collect as much data as possible before modeling.
Descriptor(Optional, applicable only to ML algorithms). Descriptors are more complex molecular representations that use specific values to represent a feature, rather than simple binary coding (molecular fingerprints). When modeling, users should select appropriate descriptors to represent molecular related information according to their research purpose.
Algorithm. ML/DL-based algorithms have significant advantages over traditional modeling methods, but they also have certain drawbacks, such as having many parameters that are difficult to tune, requiring multiple rounds of parameter tuning to build a well-performing model. To simplify the process of algorithm parameter tuning, we have exposed only key parameters and provided default parameter values, allowing users to quickly get started and build high-performing QSAR models.
The Inno-QSAR module provides the ability to quickly build AI models based on proprietary data. It supports mainstream data cleaning, data splitting, descriptor calculation, and ML/DL algorithm selection. Users can choose between automated modeling mode or manual parameter adjustment mode to obtain the best model. The results page also provides comprehensive model evaluation and statistical chart display functions, and the built model can be easily integrated into the system workflow.
2. Instructions for Use
Based on the characteristics of the QSAR model, we have divided the model construction into the following steps: selecting the modeling type - uploading the training set - uploading the test set (Optional field) - confirming the modeling data - obtaining the test set (Optional field) -selecting the modeling method.
(1) Modeling Type
The current system only supports the construction of binary classification models and regression models. You can choose the type of modeling according to your needs.
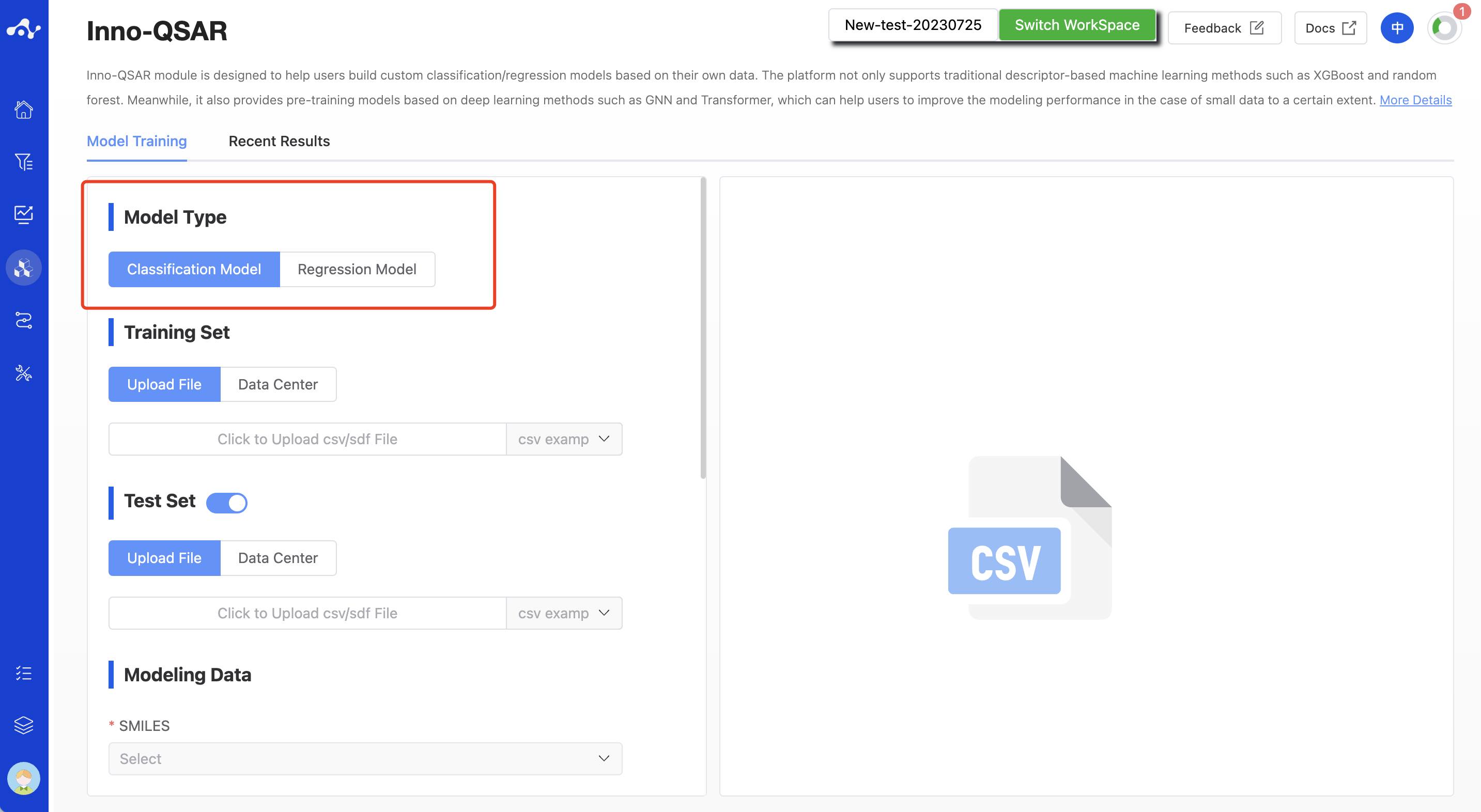
Figure 1. QSAR Modeling Page - Select Modeling Type
(2) Training Set
The training set refers to a set of compound data with known pharmacological activities. These data not only characterize the structural features of the compounds, but also contain the biological activity information of the compounds, such as IC50, EC50, etc. Currently, the platform provides upload file and data center two methods to select the training set, and supports the upload of .csv/.sdf files, which must contain SMILES and activity labels. After uploading a file, users can view the file contents in the Data Viewer on the right side.
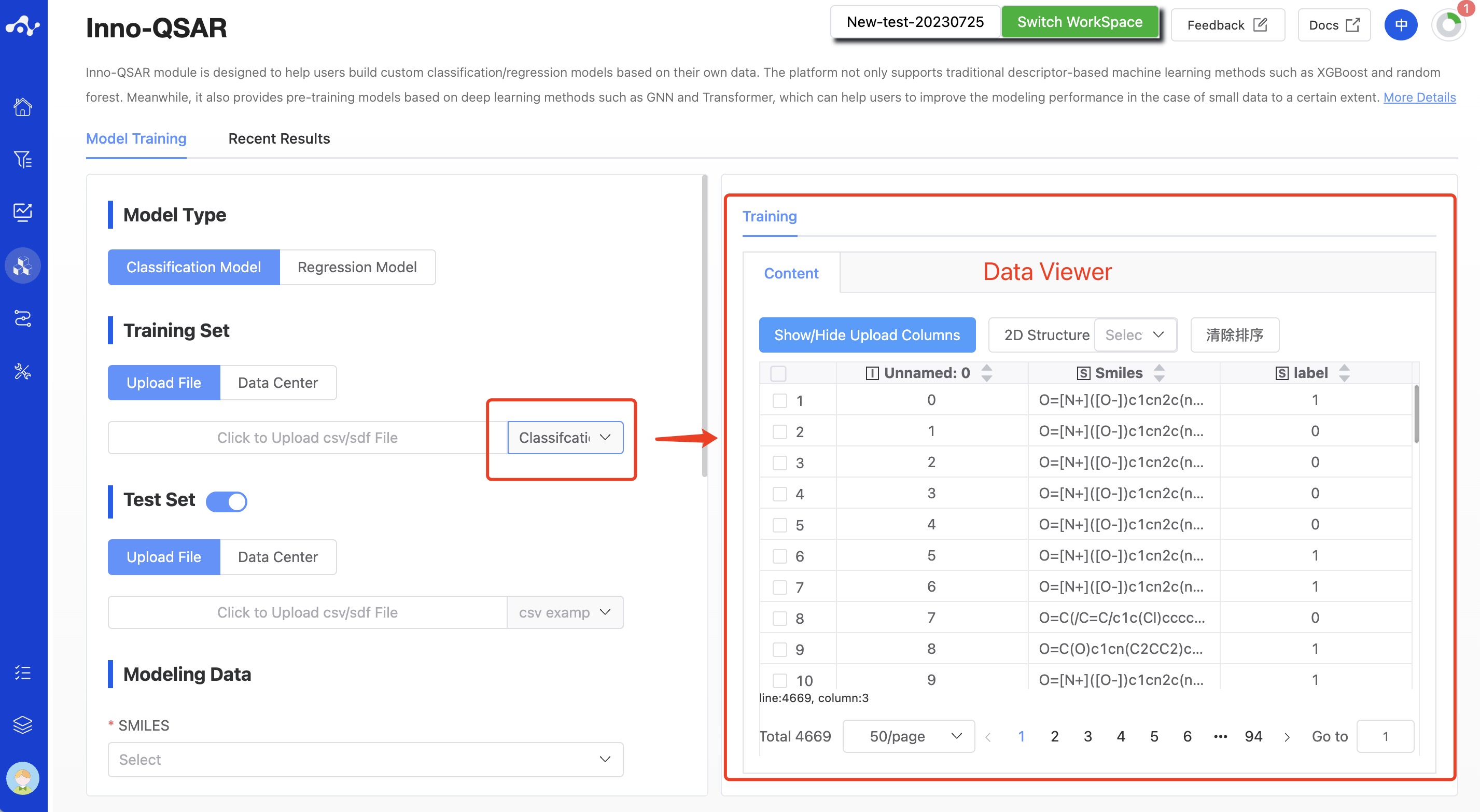
Figure 2. QSAR Modeling Page - Upload Training Set
(3) Test Set(Optional field)
The test set is an important tool for evaluating and selecting models, and plays a significant role in the establishment and optimization of the model. The system defaults to upload the test set. For users who have prepared the test set in advance, they can directly upload the test set file here.The platform provides upload file and data center two methods to select the training set, and supports the upload of .csv/.sdf files, which must contain SMILES and activity labels.
Warning: To facilitate subsequent calculations, please ensure that the SMILES names and Label names in multiple files you upload are consistent!
For users who have not prepared a test set, they can switch off this option, and then the step of "obtaining the test set" will be added later.
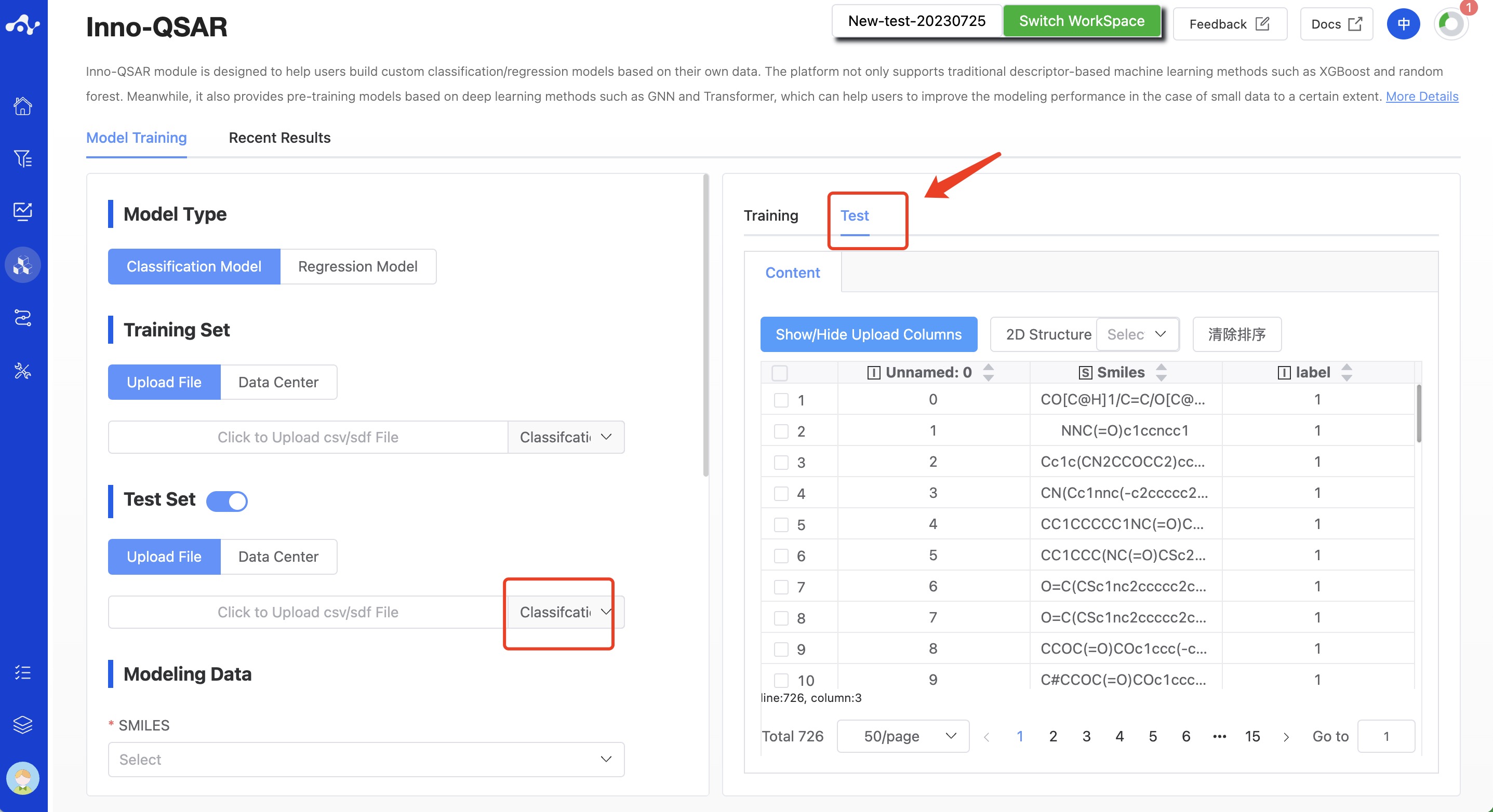
Figure 3. QSAR Modeling Page - Upload Test Set
(4) Modeling Data
After uploading the data, to build the model, we also need to specify the SMILES column, label column, and activity value column.
SMILES: The currently supported file formats are .sdf and .csv. When uploading a csv file, you need to specify the SMILES column. However, when uploading an sdf file, it's not necessary to specify the SMILES column.
Label column: Typically, this column represents activity values, such as IC50, EC50, etc. Since the current platform only supports the construction of binary classification models, the label column of classification models includes two unique values, generally "1 and 0", "Positive and Negative", "Active and Inactive". Whereas, the label column of regression models would be a series of continuous values.
Activity value column: This value is unique to classification models, used to specify the category with activity, such as "1", "Positive" or "Active".
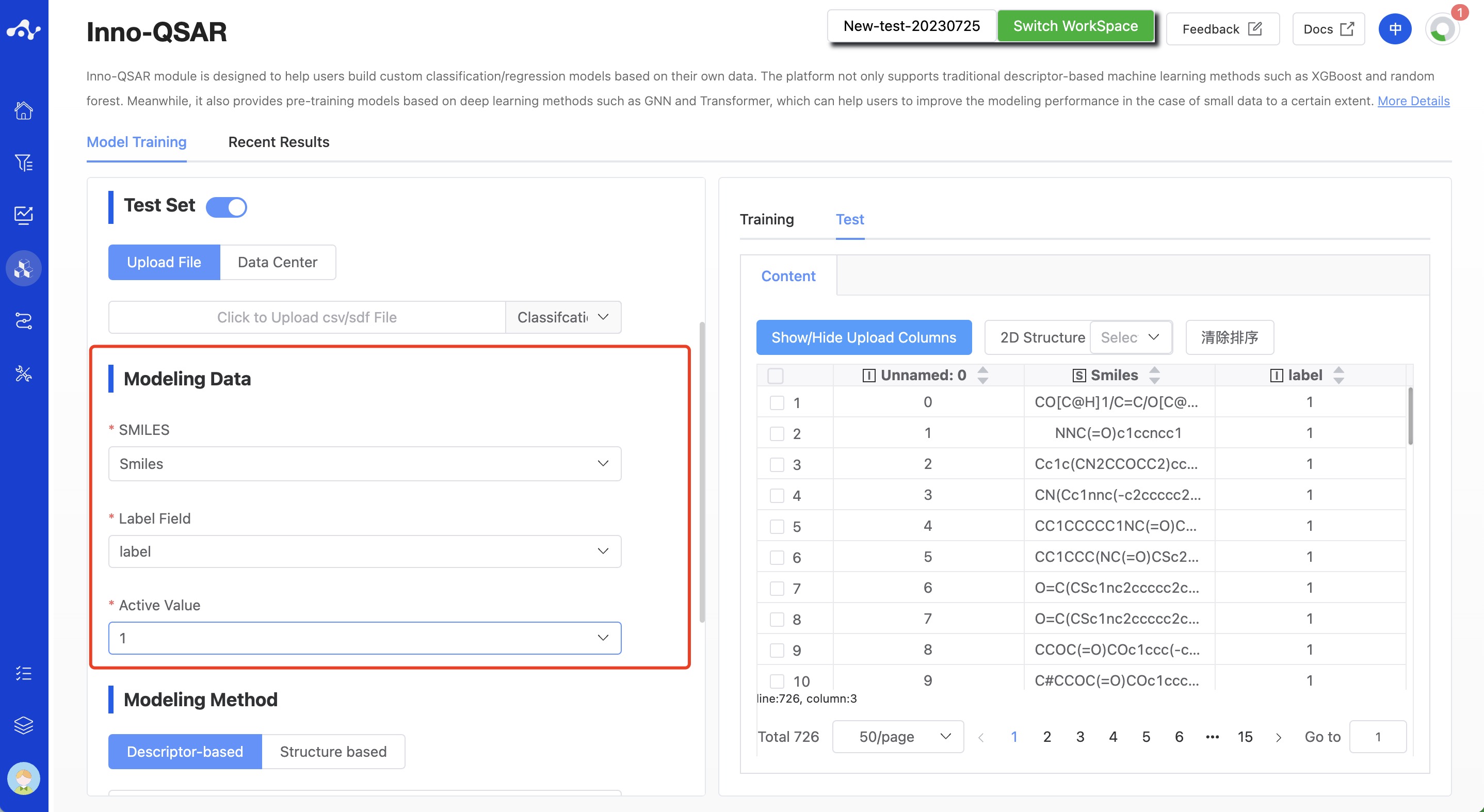
Figure 4. QSAR Modeling Page - Specify Modeling Data
(5) Obtaining the Test Set
When you switch off the entry at the upload test set step, you will be prompted to get your Test Set according to the following parameters. The default Test Set will account for 10% of the user's uploaded data and provides three sampling methods: random sampling, stratified sampling based on Label, and stratified sampling based on Murcko Scaffold.

Figure 5. QSAR Modeling Page - Obtaining the Test Set(Switch off the Test Set)
(6) Modeling Method
The platform currently provides two different classes of methods, descriptor-based and structure-based methods. Descriptor-based algorithms include AutoML, XGBoost, and RF; Structure-based algorithms include Transformer(Pretrain) and GNN(Pretrain).
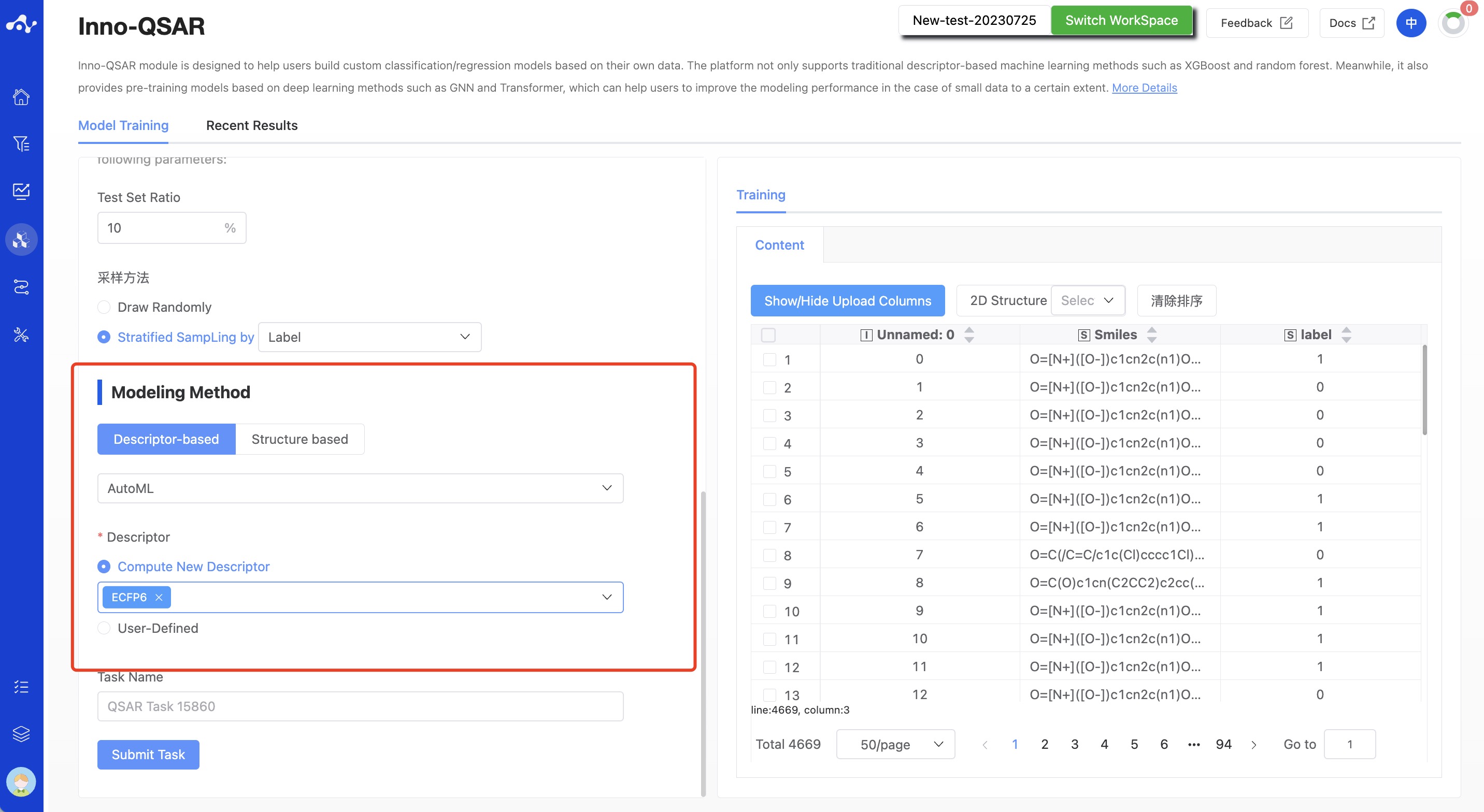
Figure 6. QSAR Modeling Page - Select Modeling Method
- Descriptor-based Method
AutoML. AutoML includes 11 algorithms. Based on the data uploaded by the user, some automated processes are used to replace traditional time-consuming manual processing steps, including handling missing data, manual feature conversion, data splitting, model selection, algorithm selection, hyperparameter selection and tuning, and ensembling multiple models. The ensembling technology of L-layer stacking & n-repeated k-bagging is applied to improve the computational speed and the accuracy of the model greatly. AutoML will adaptively remove some unsuitable models based on the input data, and then weight them based on the constructed models. The final output of AutoML is the weighted average of the successfully executed algorithm models.
The 11 algorithms mentioned above are: Random Forest, Extremely Randomized Trees, K-Nearest Neighbors, LightGBM Boosted Trees, CatBoost Boosted Trees, XGBoost Model, Linear Regression, PyTorch neural network models, VowpalWabbitModel, Fastai v1 neural network models, AutoGluon Deep Neural Networks
XGBoost. XGBoost is a machine learning algorithm based on ensemble learning, which mainly uses Gradient Boosting to build an ensemble model. Through multiple iterations, XGBoost stacks the weak classifiers so that each iteration can better fit the residual. Additionally, XGBoost uses regularization to control the complexity of the model and avoid overfitting. During model training, the combination method of grid search + five-fold cross validation is used to determine the optimal parameters, so as to improve the accuracy and generalization ability of the model and avoid performance bias due to uneven data distribution.
Random Forest. Random Forest(RF) is a decision tree-based machine learning algorithm that builds multiple decision trees by performing bootstrap sampling on the dataset and randomly selecting features multiple times for each sample. Finally, these decision trees are combined (voting or averaging) to predict new data. RF can effectively handle high-dimensional data and nonlinear relationships, while also mitigating the risk of overfitting. During model training, the combination method of grid search + five-fold cross validation is used to determine the optimal parameters, so as to improve the accuracy and generalization ability of the model and avoid performance bias due to uneven data distribution.
If users choose the Descriptor-based method, users can choose to build the model using either platform-provided descriptors (Compute New Descriptor) or User-Defined descriptors.
ECFP4/6. Extended Connectivity Fingerprints(ECFP) is a circular topology fingerprint that contain highly specific atomic information and can be used to represent substructure features. The number after ECFP represents the maximum diameter of the circle, which specifies the maximum circular neighborhood to be considered for each atom. The default diameter is 4, which can meet the requirements of similarity search and clustering, and 6/8 is favorable for large structural fragments and more suitable for activity prediction.
MACCS. MACCS descriptor is a structure descriptor that contains 166 of the most common substructure features.
Molecule Discriptors. Refers to 2D descriptors in RDKit, including 115 2D descriptors.
- Structure-based Methods
Transformer(Pretrain). Transformer(Pretrain) is a pretrained model independently developed by CarbonSilicon AI. Based on 3.3 million small molecule data, taking 1D, 2D and 3D information of molecules as input, adopting the idea of "modal alignment first, then fusion", introducing the strategies of Contrast Loss, Matching Loss and Momentum Encoder to constrain the global features of molecules, and using homo-lumo-gap acts as a supervision signal to constrain the physical properties of molecules, and using 3D molecular conformation denoising task to constrain the 3D structure of molecules, and finally applying a surrogate task of molecular fragment localization to constrain the local information of molecules. The MOE(Mixture-of-Experts) structure greatly enhances the adaptation ability of multi-modal information of the model. Verified by multiple data sets, the results show that the pre-trained Transformer model can significantly improve the modeling effect of small data sets.
GNN(Pretrain). GNN(Pretrain) is a pretrained model independently developed by CarbonSilicon AI. It takes the molecular graph encoded with atom and bond features (including 9 kinds of atom features and 5 kinds of bond features to characterize atoms, chemical bonds and their local environments) as input, extracts general features from substructures through multiple shared parameter RGCN layers, and applies attention layers to assign different attention weights to different substructures. Its advantages are: 1) the regularization effect, which enables the model to learn more tasks with the same number of parameters; 2) transfer learning effect: ADMET data is correlated, and related tasks share part of hidden layer parameters to help extract common features; 3) Data augmentation effect, ADMET data is sparse, and the model trains on all properties simultaneously to avoid overfitting on small tasks.
(7) Running Sataus and View Results
After the task is submitted, the page will automatically jump to the "Recent Results" subpage of the current page. Here you can view the task running status of the current module (progress bar), and you can also view all running tasks of all modules in the "Running" dropdown box in the upper right corner. Once the task is complete, a card will pop up in the top right corner of the page to notify you that a new task has been completed. You can click the "View Results" button in the card to view the results, or click the "Result Details" button on the current "View Results" page to view the results.
Figure 7. View Results.
3. Result Analysis
The result page consists of three parts, which are data analysis, scaffold analysis and result details.
(1) Data Analysis
The statistical distribution of the preprocessed training set and test set data is plotted to help users quickly understand the distribution of the actual data entering the model.
(2) Scaffold Analysis
Based on Murcko scaffold, a scaffold analysis on the training set and the test set was conducted, and only the top 10 skeleton structures were displayed on the page.
The scaffold classification of regression model: training set, test set
The scaffold classification of classification model: Currently, the classification model does not temporarily support skeleton analysis. We will gradually improve this in the future, presenting according to 1-training set, 1-test set, 0-test set, and 0-training set.
(3) Result details
- Classification Model
According to the characteristics of the classification model, the corresponding evaluation metrics were calculated based on the test set, including Accuracy, ROC-AUC, PRC-AUC, F1-Score, Precision, and Recall. ROC curve and PR curve were also provided.
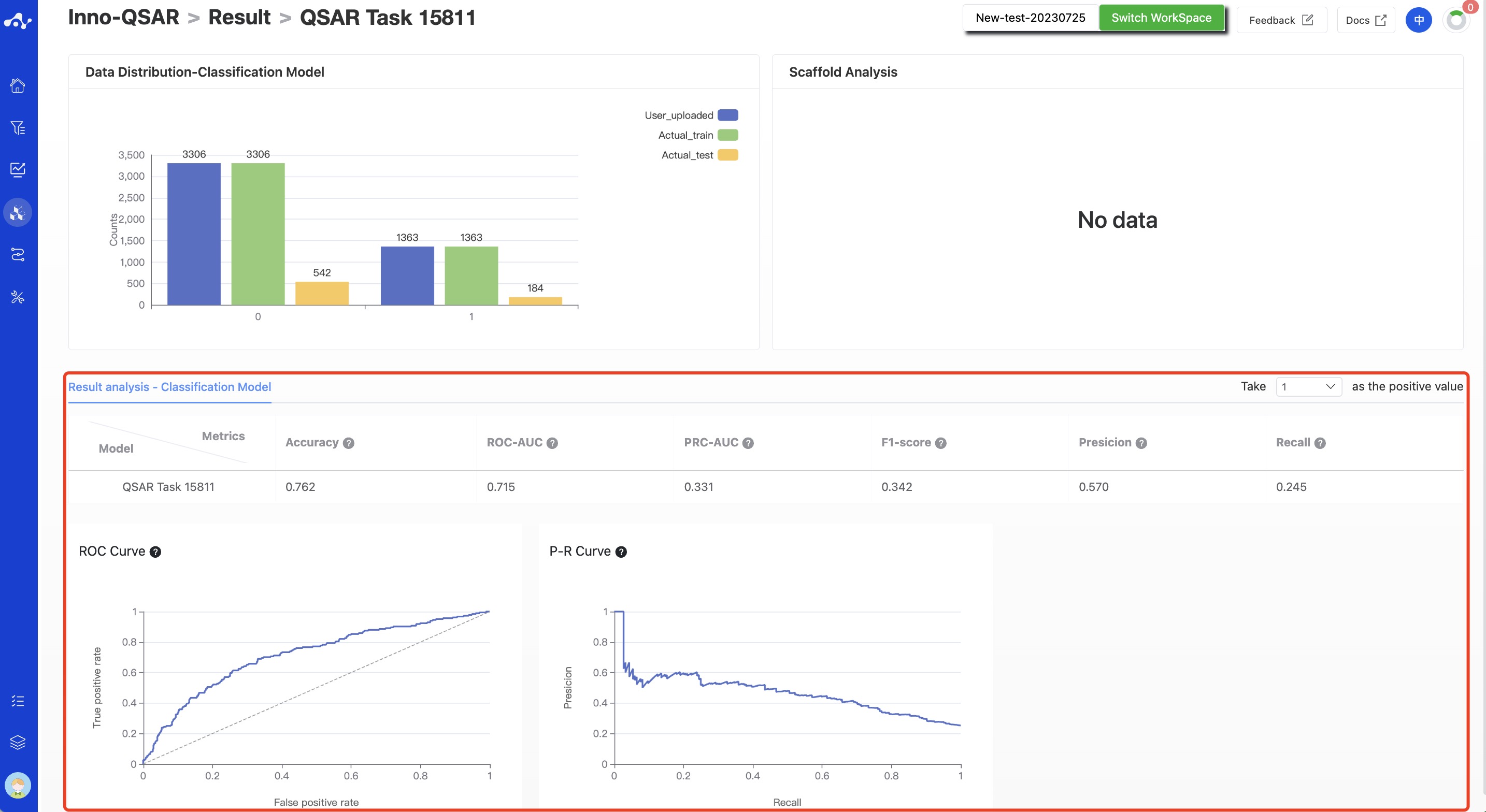
Figure 8. Result details of classification model
- Regression Model
According to the characteristics of the regression model, the corresponding evaluation metrics were calculated based on the test set, including MAE、MSE、RMSE、R2 和 PCCs,and the scatter plots of the correlation between the true values and predicted values are also provided.
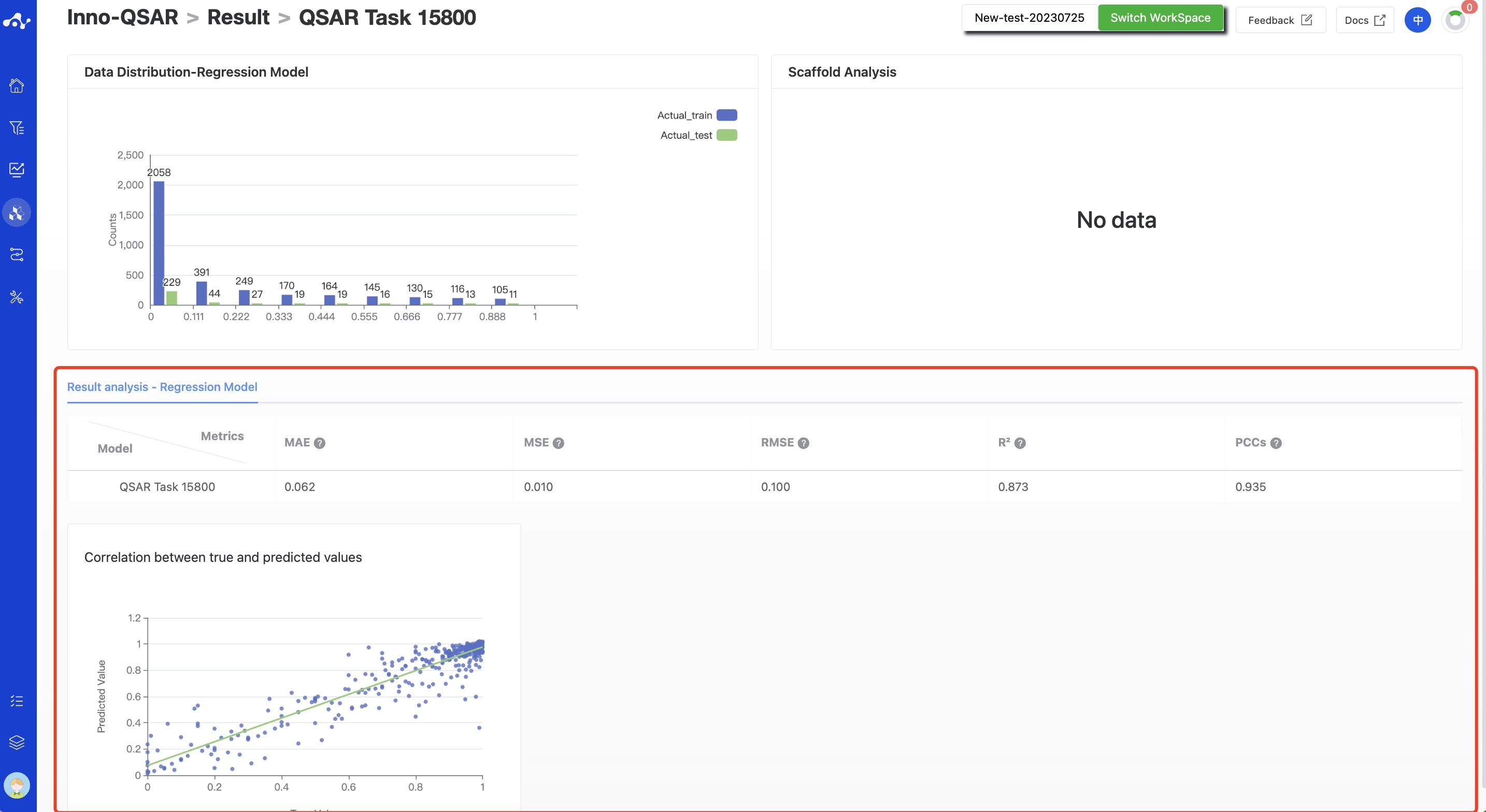
Figure 9. Result details of regression model
